Example Dataset
Fecal microbiota transplantation (FMT) is a significant breakthrough in treating recurrent Clostridium difficile infection (CDI). The authors' meticulous study reveals dynamic changes in short—and long-term bacterial composition using fecal samples from four patients before and up to 151 days after FMT, with daily collections until 28 days and weekly collections until 84 days post-FMT. The high-throughput 16S rRNA gene sequence analysis characterizes the fecal bacteria composition. FMT swiftly normalizes bacterial fecal sample composition from a markedly dysbiotic state to one representative of normal fecal microbiota.
Key points
The example dataset was from the article "Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection" (Weingarden et al. 2015). In this example dataset, we only use a subset of the complete article as described below.
1. Technology: 16S rRNA gene high-throughput sequencing was performed on fecal samples.
2. Experimental groups:
- 4x Healthy donors
- 4x Patients before fecal microbiota transplantation (FMT) to treat refractory Clostridium difficile infection.
Files
The example dataset is composed of three files that contain the eight samples:
i) The QIIME 2 artifacts for the frequency table (table.qza),
ii) The taxa composition (taxa.qza), and
iii) The metadata.csv file (metadata.csv) to tag healthy-donor and pre-FMT samples as depicted in the following figure. Remember the two mandatory columns are "sample_id" and "group." Additional columns can also be included.
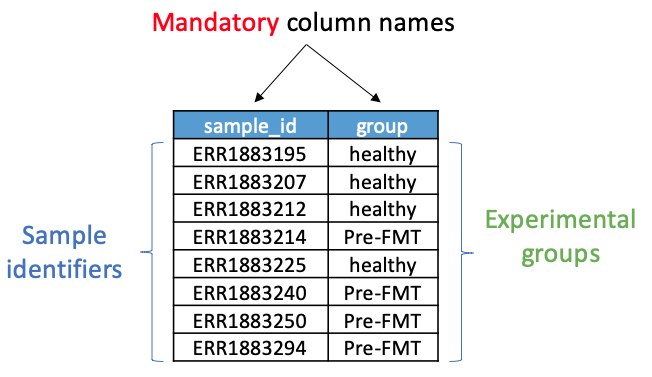
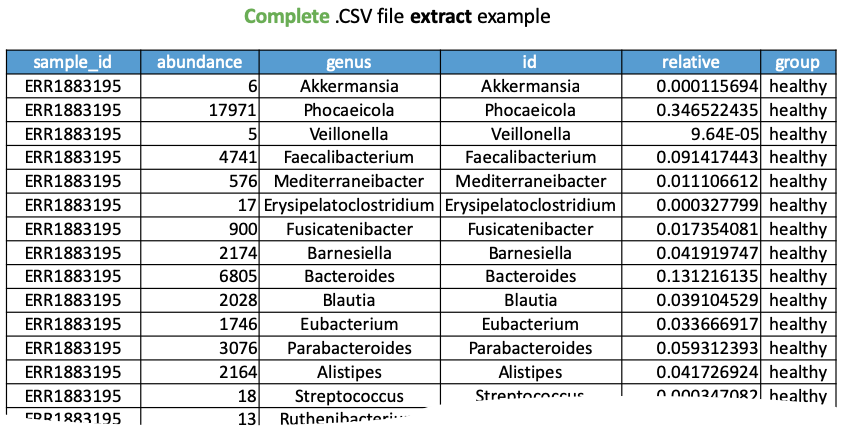
or you can download a single complete.csv file, as shown in the following Figure.
Reference
Weingarden A, González A, Vázquez-Baeza Y, Weiss S, Humphry G, Berg-Lyons D, ... & Sadowsky MJ (2015). Dynamic changes in short-and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome, 3, 1-8. https://doi.org/10.1186/s40168-015-0070-0





